Contents
How Metagenomics is Revolutionizing our Understanding of Microbial Life
Prof. Aécio D’Silva, Ph.D
AquaUniversity
What is Metagenomics?
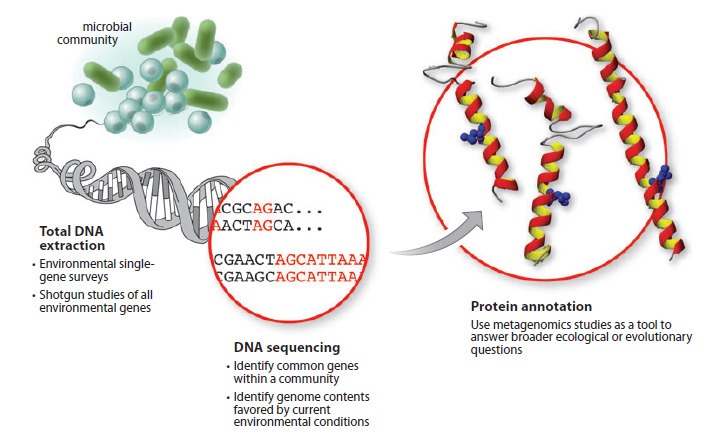
Metagenomics is a field of study that uses advanced genetic sequencing and computational techniques to analyze the collective DNA and RNA of entire microbial communities. By analyzing the genetic material of all the microorganisms in a sample, we can gain insights into the functions, interactions, and diversity of these complex ecosystems.
Metagenomics is a rapidly growing field that allows us to study the collective genetic material of entire microbial communities. By analyzing the DNA and RNA of all the microorganisms in a sample, we can gain new insights into microbial life’s complex interactions and dynamics.
Microorganisms are all around us, and they play critical roles in maintaining the health and balance of our ecosystems. However, most of these tiny organisms are difficult to study and understand because they are invisible to the naked eye and can be incredibly diverse and complex. That’s where metagenomics comes in.
Metagenomics Explained – How It Works?
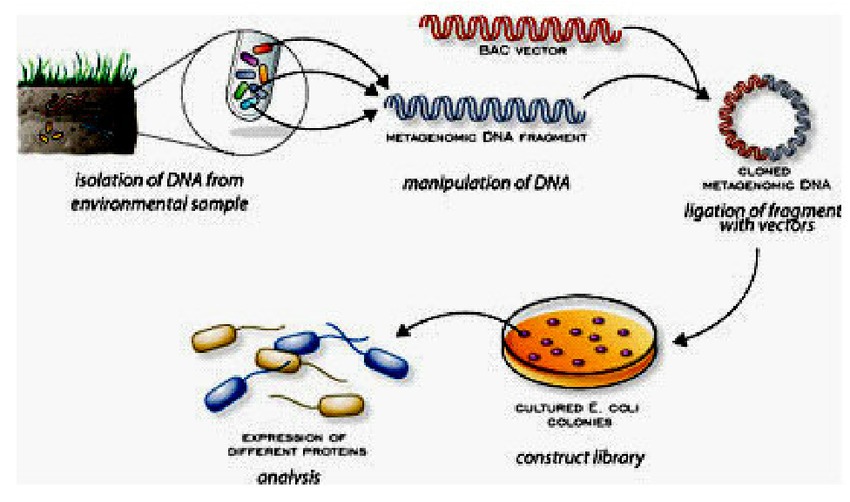
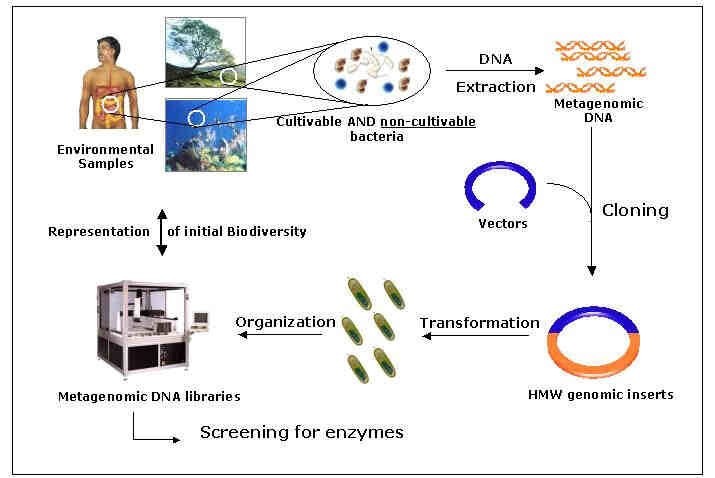
Metagenomics begins with the collection of a sample, which can come from a variety of sources, such as soil, water, or the human gut. The sample is then processed to extract genetic material, which is usually in the form of DNA or RNA. Next, the genetic material is sequenced, which means that the order of the nucleotides (the building blocks of DNA and RNA) is determined. This generates a massive amount of data that can be analyzed using powerful computational or bioinformatics analysis tools.
One of the main challenges of metagenomics is that the genetic material in a sample comes from a diverse array of microorganisms, which can be difficult to identify and classify. To address this challenge, researchers use a variety of techniques, such as comparing the genetic material to existing databases or clustering similar sequences together.
Metagenomics Explained – Applications
Metagenomics is a powerful tool for environmental microbiology. It can be used to study the biodiversity of microbes in soil, water, and air samples.
Metagenomic sequencing has also been applied to human microbiome studies where the goal is to identify all of the microorganisms that live on or inside people’s bodies. This information can help researchers understand how these organisms interact with each other and their hosts, as well as why certain diseases occur more frequently than others in certain populations (e.g., obesity).
In reality, Metagenomics has a wide range of applications in fields such as environmental science, agriculture, medicine, and biotechnology. For example, in environmental science, metagenomics can be used to study the diversity and function of microbial communities in different ecosystems, such as the ocean, soil, or the human gut. This information can be used to better understand the role of microorganisms in maintaining ecosystem health and resilience.
In agriculture, metagenomics can be used to study the microbial communities that live on and in crops, which can significantly impact plant health and productivity. By understanding the functions and interactions of these microbial communities, researchers can develop strategies to improve crop yield and reduce the use of pesticides and fertilizers.
In aquaculture, we’ve utilized metagenomics to study AquaBioPonics Bioflocs system, which synergistically combines aquaculture with bioflocs technology, algae cultivation, hydroponics, and rainwater harvesting, is an efficient aquaculture biotechnology to produce fish, shrimp, and organic vegetables. By adding a commercial probiotic to our systems, we observed improved productivity and significantly lower ammonia concentrations. Our metagenomics research gave valuable insight into the overall microbiome and the presence of nitrogen-metabolizing bacteria present in these high-yield aquaculture systems.
In medicine, metagenomics is being used to study the microbiome, which is the collection of microorganisms that live in and on the human body. By analyzing the genetic material of these microorganisms, researchers can gain insights into the role of the microbiome in health and disease. This information can be used to develop new treatments and therapies for a wide range of conditions, such as inflammatory bowel disease, obesity, and cancer.
Metagenomics Explained – Challenges and Future Directions
Despite its many applications, metagenomics still faces several challenges. One of the biggest challenges is the vast amount of data that is generated by sequencing the genetic material of entire microbial communities. This requires advanced computational tools and expertise to process and analyze the data.
Another challenge is the need for better methods to identify and classify the microorganisms and excellent throughput sequencing machines require standard computing tools and software with excellent data analysis accuracy in order to analyze the sequencing data of viral genomes. High-cost technical engagement is required for this.
To conclude, Metagenomics is a powerful tool for studying and understanding the genome of an organism. It has many advantages, including:
The ability to analyze samples from multiple individuals at once. This allows for comparison between different populations, as well as within a single population over time or space.
A wide range of applications in medicine, agriculture, aquaculture, and environmental sciences (among others). However, there are also some limitations to metagenomics that must be considered when designing experiments. However, its potential is immense, and we are only at the beginning of the pioneering of this incredible technology.
References:
- (1) Aecio D’Silva and John A Kyndt (2020) Bacterial Diversity Greatly Affects Ammonia and Overall Nitrogen Levels in
Aquabioponics Bioflocs Systems, Based on 16S rRNA Gene Amplicon Metagenomics. Applied Microbiol Open Access 6: 169. Doi: 10.35248 / 2471-9315.20.6.169 - (2) Bhukya PL and Nawadkar R (2018) Potential Applications and Challenges of Metagenomics in Human Viral Infections. Metagenomics for Gut Microbes. InTech. DOI: 10.5772/intechopen.75023.