Metagenomics Analysis – Bacterial Diversity Greatly Affects Ammonia and Overall Nitrogen Levels in Aquabioponics Bioflocs Systems, Based on 16S rRNA Gene Amplicon Metagenomics**
Aecio D’Silva1, John A Kyndt2*
1Department Moura Enterprises, Aquaculture Biotechnology Division, Tucson, AZ 85742, USA; 2Department College of Science and Technology, Bellevue University, Bellevue, NE 68005, USA
**This article was originally published at: Applied Microbiology: Open Access ISSN: 2471-9315
ABSTRACT
Metagenomics Analysis – AquaBioPonics Bioflocs system, that synergistically combines aquaculture with bioflocs technology, algae cultivation, hydroponics, and water maximization use, is efficient aquaculture biotechnology to produce fish, shrimp, and organic vegetables. By adding a commercial probiotic to our systems, we observed improved productivity and significantly lower ammonia concentrations. We provide initial insight into the overall microbiome and the presence of nitrogen-metabolizing bacteria present in these high-yield aquaculture systems.
Keywords: Aquaculture; Metagenomics; Comammox bacteria; Nitrospira; Probiotics; AquaBioPonics Bioflocs
INTRODUCTION
Metagenomics Analysis – The efficiency of AquaBioPonics systems can be strongly improved when bioflocs technology (BFT) is added as water quality management technique. Bioflocs by their definition, are clustered aggregations of microbial communities such as phytoplankton, zooplankton, bacteria, and living and dead particulate organic matter. Shrimp and tilapia especially benefit from BFT due to their ability to filter-feed on floc in the water column, thereby reducing feed costs by improving feed conversion [1,2]. Recently, studies also show that bioflocs can be collected from culture systems, dried and added as a component to pelleted aquaculture feeds, substituting 2/3 of fish meal and 100 percent of plant meals. However, the economic feasibility of employing flocs as a dried feed constituent demands more research [3]. To promote biofloc production, it is important to control the pH (7-8), ammonia, nitrite, nitrate, and sludge of the tanks/raceways. In biofloc systems, there are three main processes that control ammonia: algal uptake, bacterial assimilation, and nitrification [4]. To maximize these processes, balancing input C:N ratio is the major management strategy for ammonia control in bioflocs systems, however it is expected that the bacterial diversity of the aquaculture plays an important role as well. To study the bacterial diversity, we tested our AquaBioPonic Bioflocs systems with probiotics (APB; Aquabioponics ProBiotics) and without applying commercial probiotics (ANPB; Aquabioponics Non-ProBiotics). These probiotic formulas are designed to digests organic sludge, improve FCR and reduce ammonia, nitrite, and nitrate. In this report, we describe how the ammonium concentration in the cultures was reduced to very low levels, indicating the ammonium was transformed into microbial biomass. To understand more about the microbial processes involved in this, we compared the microbial composition using 16S RNA amplicon metagenomics analysis between tanks with (APB) and without probiotics additions (ANPB).
MATERIALS AND METHODS
Metagenomics Analysis – The experiments ran for 60 days at ACI FishFarm, Florence Arizona, using 6 nursery circular tanks (10 cubic meters –12,000 fish with an initial average weight of 0.9 g – final average weight 32.7 g) and 6 raceways (201 cubic meters with 12,000 fish with an initial average weight of 36.5 g – final average weight 153.5 g), comparing them with other tanks/raceways also in full production, but not using the probiotic (Figure 1). Fish (Tilapia) were growing for 15 days in the circular tanks and 35 days in the raceways, before applying the probiotics in a final concentration of 0.5 mg/L. Total ammonium concentrations (expressed as NH3-N), nitrite and nitrate concentrations were measured using the Total Nitrogen, Nitrite, and Nitrate Reagent Hach Kit (Hach).
Researchers Information:
Correspondence to: John A Kyndt, Department College of Science and Technology, Bellevue University, Bellevue, NE 68005, USA, E-mail: jkyndt@bellevue.edu or Aecio D’Silva. Moura Biotechnology Division, USA, E-mail: support@mybelojardim.com
Received: June 20, 2020; Accepted: July 4, 2020; Published: July 10, 2020
Citation: D’Silva A, Kyndt JA (2020) Bacterial Diversity Greatly Affects Ammonia and Overall Nitrogen Levels in Aquabioponics Bioflocs Systems, Based on 16S rRNA Gene Amplicon Metagenomics. Appli Microbiol Open Access 6:169. Doi: 10.35248/2471-9315.20.6.169
Copyright: © 2020 D’Silva A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Figure 1: AquaBioPonics Bioflocs systems at ACI FishFarm, Florence Arizona, are set up as open, recirculating raceway (top images) or nursery circular tanks (bottom images).
Water samples for 16S rRNA metagenomics analysis were collected in September 2019 from our tanks and raceways, shipped overnight to the lab and stored at 4°C. Samples were taken from both the ANPB and APB tanks 10 days after the addition of the probiotics to the APB tanks. Samples were collected in triplicate and pooled for each condition. Total DNA was extracted using the PureLink Microbiome DNA Purification Kit (Invitrogen), resulting in an absorbance 260/280 ratio of 1.98 (ANPB) and 1.92 (APB) as measured by Nanodrop (Thermo Scientific). A 16S rRNA amplicon sequencing library was prepared for each sample, following the 16S Metagenomic Sequencing Library Preparation protocol (Illumina), using amplicon primers targeting the V3 and V4 regions [5]. The samples were sequenced using a 1.8 pM library with an Illumina MiniSeq using paired-end (2 ×150 bp) sequencing. Primer sequences were removed and reads with low-quality scores (average score<20) were filtered out using the FASTQ Toolkit (Illumina, version 2.2.0). The 16S Metagenomics app within BaseSpace (version 1.0.1) was used to perform a taxonomic classification, which uses an Illuminacurated version of the GreenGenes taxonomic database [6]. Default parameters were used for all the software applications.
RESULTS AND DISCUSSION
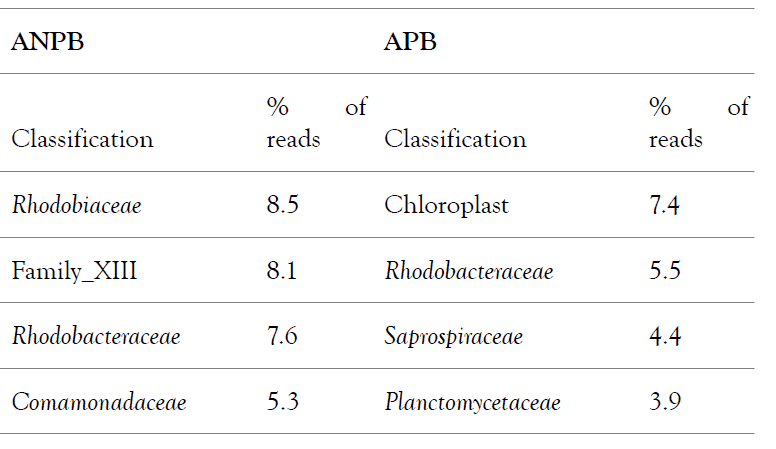
Metagenomics Analysis – After 10-20 days, the total ammonium concentration in the tanks and raceways were steadily at very low levels (=< 1.0 ppm) in the probiotic-added tanks (APB), indicating that ammonium was transformed into microbial biomass. Nitrite, and nitrate, and sludge are also all within safe levels. Before we started using probiotics and in the ANPB tanks raceways/tanks, these levels reached 8.0 ppm on average and frequently spiked higher. To control these parameters, we previously had to constantly apply a locally made bioflocs mix (brown/white sugars, molasses, starch, cassava, and flours – wheat, oat, corn). This substantially increased the labor intensity to maintain good water quality levels. The basic differences observed in fish (Tilapia) yield were increased survival rates (between 95% and 99%) and feed conversation ratios (=< 1.0). We harvested the fish in the tanks (at the end of the 60-day experiment) and continued in the raceways until the fish reached 850 g (total time: 135 days). Total DNA was extracted from water samples (5 ml) taken from both the tanks and the raceways, obtaining an average yield of 5 ng/ul for the ANPB samples and 50 ng/ul for the APB samples, with a 260/280 ratio of 1.98 (ANPB) and 1.92 (APB). Illumina sequencing generated 438,278 reads for ANPB and 437,626 reads for APB, which were analyzed for metagenomic composition using the Metagenomics app within Basespace (Illumina). The two samples showed a clear difference in bacterial composition at the family level and beyond. Table 1 illustrates the most abundant OTUs showing a significant difference at the family level between the two samples. 79% of the reads were classified to the genus level.
Table 1: Overview of the bacterial diversity at the family level, based on 16S rRNA gene amplicon metagenomics analysis of samples from Aquabioponics systems. ANPB is a sample without probiotics added, while APB is a probiotic containing the sample. Only families with abundances over 3.5% of the reads are represented.
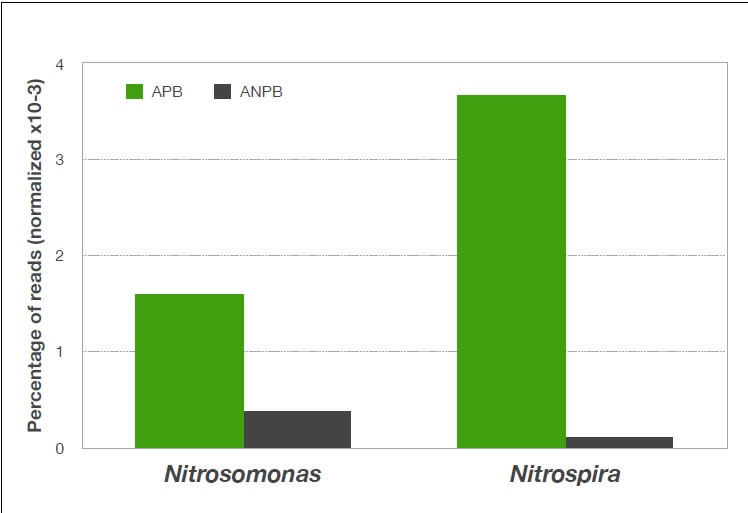
Notably, there was no chloroplast representation in the ANPB top results, indicating less algal growth in these aquacultures. According to a Shannon Species Diversity Analysis [7,8] each of the samples contained around 2000 potential species. There appears to be a substantially higher representation of nitrogen metabolizing species in the APB culture compared to the ANPB culture (Figure 2). Nitrosomonas contained 700 reads in APB and 167 reads in ANPB, and Nitrospira contained 1608 reads in APB versus only 48 reads in ANPB species. No significant amounts of Nitrobacter species were found in either sample. These results are consistent with our observation of significantly less ammonia and higher nitrogen assimilation in our APB aquacultures.
Figure 2: Chart overview of the nitrogen-metabolizing species in the APB cultures compared to the ANPB cultures. Values are presented as the percentage of reads for each species, normalized to the total number of reads for each sample.
Several Nitrospira isolates are considered comammox (COMplete AMMonia OXidiser) bacteria [9,10] and these appear to be the major nitrogen-metabolizing species in the system. Based on our 16SrRNA identification, Nitrospira japonica (NR_114396.1) was the dominant Nitrospira species in our cultures, while Nitrosomonas aestuarii (FR828476), Nitrosomonas oligotropha (FR828478), and Nitrosomonas ureae (FR828472) were the main Nitrosomonas species. Nitrification is a central part of the nitrogen cycle, and for more than a century, nitrification was thought to be a two-step process involving ammonia oxidation to nitrite by chemolithoautotrophic ammonia-oxidizing microorganisms and nitrite oxidation to nitrate by nitrite-oxidizing bacteria. All known comammox are members of the genus Nitrospira and possess complementary enzymes for both functions of ammonia and nitrite oxidation [9,10]. Metagenomic evidence of the presence of comammox in drinking water distribution systems [11, 12], groundwater-fed rapid sand filters [13,14], wastewater treatment processes [15-18], and agriculture soils [19] has been reported. Species identification based on only 16S rRNA analysis is certainly limiting, and it has been shown that 16S rRNA alone should not be used to identify comammox at the species level because these bacteria are not distinguishable via phylogenetic analyses from strict nitrite-oxidizing bacteria in the genus Nitrospira. Further analysis would be needed to identify the true species identity of the dominant uncultured Nitrospira species in our aquabioponics systems.
CONCLUSION
AquaBioPonics Bioflocs have been shown to be highly efficient, especially for high-density tilapia aquaculture, and based on this analysis we can conclude that the microbial composition, particularly comammox bacteria, appears to play an important role in removing the excess nitrogen that will otherwise poison the fish. This method not only improves the water nitrogen quality but also significantly reduces the labor involved in maintaining these systems.
Data availability
The 16S rRNA gene amplicon data sets have been deposited at DDBJ/ENA/GenBank under project PRJNA599405 and can be accessed with the SRA accession numbers SRR10843757 (ANPB) and SRR10843772 (APB).
ACKNOWLEDGEMENTS
This work was sponsored by the Wilson Enhancement Fund for Applied Research in Science at Bellevue University. Arizona Correctional Industries Complex FishFarm at Florence, Arizona provided the facility to run the experiments. Dr. Marcos Eberlin from Mackenzie Presbyterian University, São Paulo, Brazil, for introducing the authors to Nitrospira species.[20]
REFERENCES
1. McGraw B. Biofloc systems viable for tilapia production. Global Aquaculture Alliance. 2019;
2. Crab R, Defoirdt T, Bossier P, Verstraete W. Biofloc technology in aquaculture: Beneficial effects and future challenges. Aquaculture. 2012; 351-356.
3. Hargreaves JA. Biofloc Production Systems for Aquaculture. SRAC Publication. No. 4503 2013;
4. Klindworth A, Pruesse E, Schweer, T, Peplies, J, Quast, C, Horn, M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013; 41:1: e1.
5. Wang Q, Garrity, GM, Tiedje, JM, Cole, JR. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy, Appl. Env. Microbiol. 2007; Vol. 73. 16: 5261–5267.
6. Spellerberg IF, and Peter JF. A tribute to Claude Shannon and a plea for more rigorous use of species richness, species diversity, and the ‘Shannon–Wiener’ Index. Global Ecology and Biogeography. 2003; 12:3: 177-179.
7. Tuomisto H. A diversity of beta diversities: straightening up a concept gone awry. Part 1. Defining beta diversity as a function of alpha and gamma diversity. Ecography. 2010; 33: 2–22.
8. Daims H, Lebedeva E V, Pjevac P, Han P, Herbold C, Albertsen M, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015; 528: 504–509.
9. Van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, et al. Complete nitrification by a single microorganism. Nature. 2015; 528: 555–559.
10. Pinto AJ, Marcus DN, Ijaz UZ, Santos QMBD, Dick GJ, Raskin L. Metagenomic evidence for the presence of comammox Nitrospiralike bacteria in a drinking water system. mSphere. 2016;1:e00054-15.
11. Wang YL, Ma LP, Mao YP, Jiang XT, Xia Y, Yu K, et al. Comammox in drinking water systems. Water Res. 2017; 116:332–341.
12. Bartelme RP, McLellan SL, Newton RJ. Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing archaea and comammox Nitrospira. Front Microbiol. 2017; 8:101.
13. Fowler SJ, Palomo A, Dechesne A, Mines PD, Smets BF. Comammox Nitrospira are abundant ammonia oxidizers in diverse groundwater-fed rapid sand filter communities. Environ. Microbiol. 2018; 20:1002–1015.
14. Annavajhala MK, Kapoor V, Santo-Domingo J, Chandran K. Comammox functionality identified in diverse engineered biological wastewater treatment systems. Environ. Sci. Technol. Lett. 2018; 5:110–116.
15. Camejo PY, Domingo JS, McMahon KD, Noguera DR. Genomeenabled insights into the ecophysiology of the comammox bacterium “ Candidatus Nitrospira nitrosa. ” mSystems. 2017;
2:e00059-17.
16. Fan XY, Gao JF, Pan KL, Li DC, Dai HH. Temporal dynamics of bacterial communities and predicted nitrogen metabolism genes in a full-scale wastewater treatment plant. RSC Adv. 2017; 7:56317–56327.
17. Chao Y, Mao Y, Yu K, Zhang T. Novel nitrifiers and comammox in a full-scale hybrid biofilm and activated sludge reactor revealed by metagenomic approach. Appl. Microbiol. Biotechnol. 2016;
100:8225–8237.
18. Orellana LH, Chee-Sanford JC, Sanford RA, Loffler FE, Konstantinidis KT. Year-round shotgun metagenomes reveal stable microbial communities in agricultural soils and novel ammonia oxidizers responding to fertilization. Appl. Environ. Microbiol. 2018; 84:e01646.
19. Daims H, Lucker S, Wagner M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 2016; 24 : 699–712.
20. Eberlin Marcos. Foresight: How the Chemistry of Life Reveals Planning and Purpose. 2019. Discovery Institute Press, Seattle, WA, USA.